Cooperative Isotherms via Statistical Thermodynamics
Quick Start
Cooperative isotherms, IUPAC Types IV, V & VI, can be described elegantly via the Stat Therm approach described in other apps on this site, modified to work with equations specifically suited to cooperativity.
Credits
The app is based on core papers by Shimizu and Matubayasi1-2 and a subsequent paper with co-authors Dalby and Abbott3.
Cooperative Isotherms
//One universal basic required here to get things going once loaded
window.onload = function () {
//restoreDefaultValues(); //Un-comment this if you want to start with defaults
document.getElementById('Load').addEventListener('click', clearOldName, false);
document.getElementById('Load').addEventListener('change', handleFileSelect, false);
document.getElementById('Demo').addEventListener('change', GetDemo, false);
GetDemo()
}
// Main(); //No need because GetDemo does it};
//Any global variables go here
let amUpdating = false, lastFit = "", newFit = false, lastQ = 0, lastUnits = "", lastProbe = "", lastT = 0, divBy = 1, ModelLabel = "Model Parameters", G22conv = 1
let fitData = [], naFitData = [], scaleData = [], Loading = false
//Main is hard wired as THE place to start calculating when input changes
//It does no calculations itself, it merely sets them up, sends off variables, gets results and, if necessary, plots them.
function Main() {
saveSettings();
if (Loading) return
//Send all the inputs as a structured object
//If you need to convert to, say, SI units, do it here!
const inputs = {
A:0,
B: 0,
C: 0,
D: 0,
amax: 1,
Q: sliders.SlideQ.value,
Probe: document.getElementById('Probe').value,
T: sliders.SlideT.value,
Units: document.getElementById('Units').value,
Model: document.getElementById('Model').value,
}
//Send inputs off to CalcIt where the names are instantly available
//Get all the resonses as an object, result
if (!amUpdating) console.time("Calc")
const result = CalcIt(inputs)
if (!amUpdating) console.timeEnd("Calc")
//Set all the text box outputs
// document.getElementById('Langmuir').value = result.Lang
document.getElementById('Parameters').value = result.Parameters
document.getElementById('ModelLabel').innerHTML = result.ModelLabel
//Do all relevant plots by calling plotIt - if there's no plot, nothing happens
//plotIt is part of the app infrastructure in app.new.js
if (result.plots) {
for (let i = 0; i < result.plots.length; i++) {
plotIt(result.plots[i], result.canvas[i]);
}
}
//You might have some other stuff to do here, but for most apps that's it for CalcIt!
}
//Here's the app calculation
//The inputs are just the names provided - their order in the curly brackets is unimportant!
//By convention the input values are provided with the correct units within Main
function CalcIt({ A, B, C, D, Q, amax, Model, Probe, Units, T }) {
const FQ = 2000 + 2000 * Q * Q
const ProbeData = Probe.split(";")
const MWt = parseFloat(ProbeData[1])
const Area = parseFloat(ProbeData[2])
const AA = parseFloat(ProbeData[3])
const AB = parseFloat(ProbeData[4])
const AC = parseFloat(ProbeData[5])
if (amUpdating) return { Parameters: "-" }
document.getElementById('Load').disabled = !Model.includes("File")
if (Model.includes("File")) {
document.getElementById('SlideQ').style.visibility = "visible"
} else {
document.getElementById('SlideQ').style.visibility = "hidden"
Loading = false
}
const RT = 8.314 * 298
let ICurve = [], GCurve = [], NCurve = [], anCurve = [], PCurve = [], naCurve = [], G22 = 0, n = 0, E = 0, F = 0, G = 0, H = 0
let Parameters = "-", Values = "-"
const a2inc = 0.01
if (!Model.includes("File")) {
fitData = []; naFitData = []
clearOldName()
Loading = false
}
ModelLabel = "Model Parameters"
//Handle the Files first
let xmax = amax - 0.05
if (Model.includes("File") && fitData.length > 3) {
// document.getElementById('Slideamax').style.visibility = "hidden"
let x = [], y = []
naFitData = []
for (let i = 0; i < fitData.length; i++) {
x[i] = fitData[i].x
y[i] = fitData[i].y
if (x[i] > 0.025) naFitData.push({ x: fitData[i].x, y: fitData[i].y * divBy / fitData[i].x })
}
xmax = parseFloat(x[fitData.length - 1])
if (amax < xmax) amax = Math.min(1, xmax + 0.05) // Users can choose to have a higher value but not a lower one
if (newFit || Q != lastQ || Probe != lastProbe || Units != lastUnits || lastFit != Model) {
lastFit = Model; lastQ = Q; lastUnits = Units; lastProbe = Probe
amUpdating = true
let conv = 1
if (!Units.includes("mol")) conv *= MWt
if (Units.includes("/g") & !Units.includes("mmol") & !Units.includes("μmol")) conv /= 1000
if (Units.includes("μmol")) conv /= 1000
if (Units.includes("/100g")) conv /= 10
if (Model.includes("Type IV")) {
//Although Type IV data can be fitted by the native formula
//the fitting can be re-cast using 3 parameters from the curve itself
//This is both faster and likely to have fewer fitting artefacts.
PFunc = function (x, P) {
return x.map(function (xi) {
// return P[0] * (P[1] * xi) / (1 + P[1] * xi) + P[2] * (P[3] * P[4] * Math.pow(xi, P[3])) / (1 + P[4] * Math.pow(xi, P[3]))
return P[0] * (P[1] * xi) / (1 + P[1] * xi) + P[2] * (P[3] * Math.exp(P[3] * (P[4] + Math.log(xi)))) / (1 + Math.exp(P[3] * (P[4] + Math.log(xi))))
})
}
let dmax = 0, a2mid = 0
for (i = 1; i < y.length; i++) {
tmp = (y[i] - y[i - 1]) / (x[i] - x[i - 1])
if (isFinite(tmp) && tmp > dmax) { dmax = tmp; a2mid = x[i] }
}
const h = Math.max(...y)
let xval = 1, N = 1, m = 40
//The values from the curve - assuming they make sense
if (a2mid > 0.1*xmax && a2mid < 0.9*xmax && dmax > 0) {
xval = -Math.log(a2mid)
N = h * h / (4 * dmax * a2mid)
m = h / N
}
// var Parms = fminsearch(PFunc, [0.5, 1, 1, 5, 1000], x, y, {
// maxIter: FQ, noNeg:true
// })
// A = Parms[0]; B = Parms[1]; C = Parms[2]; D = Parms[3]; E = Parms[4]
var Parms = fminsearch(PFunc, [0.5, 1, N, m, xval], x, y, {
maxIter: FQ, noNeg: true
})
//Am comes from the combination of xval and m
A = Parms[0]; B = Parms[1]; C = Parms[2]; D = Parms[3]; E = Math.exp(Parms[3] * Parms[4])
const RTlnA=-8.314*(T+273)*Math.log(E)/1000 //kJ/mole
Parameters = "Na : " + (A * divBy * conv).toExponential(2) + " mol/kg, A1 : " + B.toPrecision(3) + " Nb : " + (C * divBy * conv).toExponential(2) + " mol/kg, m : " + D.toPrecision(4) + ", Am : " + E.toPrecision(3)+ ", -RTln(Am) : "+RTlnA.toPrecision(3)+"kJ/mol"
let cText = (A * divBy * conv).toExponential(2) + '\t' + B.toPrecision(3) + '\t' + (C * divBy * conv).toExponential(4) + '\t' + D.toPrecision(4) + '\t' + E.toPrecision(3) + '\t' + RTlnA.toPrecision(3)+"kJ/mol"
copyTextToClipboard(cText)
}
if (Model.endsWith("Type V")) {
PFunc = function (x, P) {
return x.map(function (xi) {
return P[0] * (P[1] * xi + P[3] * P[2] * Math.pow(xi, P[3])) / (1 + P[1] * xi + P[2] * Math.pow(xi, P[3]))
})
}
const ymax = Math.max(...y)
var Parms = fminsearch(PFunc, [ymax / 10, 1, 100, 10], x, y, {
maxIter: FQ, noNeg: true
})
A = Parms[0]; B = Parms[1]; C = Parms[2]; D = Parms[3]; E = 0
Parameters = "N : " + (A * divBy * conv).toExponential(2) + " mol/kg, m : " + D.toPrecision(3) + ", A1 : " + B.toPrecision(3) + ", Am : " + C.toPrecision(4)
let cText = (A * divBy * conv).toExponential(2) + '\t' + D.toPrecision(3) + '\t' + B.toPrecision(3) + '\t' + C.toPrecision(4)
copyTextToClipboard(cText)
}
if (Model.endsWith("Type VI")) {
PFunc = function (x, P) {
return x.map(function (xi) {
return P[0] * (P[1] * xi) / (1 + P[1] * xi) + P[2] * (P[3] * P[4] * Math.pow(xi, P[3])) / (1 + P[4] * Math.pow(xi, P[3])) + P[5] * (P[6] * P[7] * Math.pow(xi, P[6])) / (1 + P[7] * Math.pow(xi, P[6]))
})
}
const ymax = Math.max(...y)
var Parms = fminsearch(PFunc, [0.5, 1, 0.005, 5, 1000, 0.0005, 30, 10000], x, y, {
maxIter: FQ, noNeg: true
})
A = Parms[0]; B = Parms[1]; C = Parms[2]; D = Parms[3]; E = Parms[4]; F = Parms[5]; G = Parms[6]; H = Parms[7]
Parameters = "Na : " + (A * divBy * conv).toExponential(2) + " mol/kg, A1 : " + B.toPrecision(3) + " Nb : " + (C * divBy * conv).toExponential(2) + " mol/kg, m : " + D.toPrecision(3) + ", Am : " + E.toPrecision(3) + " Nc : " + (F * divBy * conv).toExponential(2) + " mol/kg, n : " + G.toPrecision(3) + ", An : " + H.toPrecision(3)
let cText = (A * divBy * conv).toExponential(2) + '\t' + B.toPrecision(3) + '\t' + (C * divBy * conv).toExponential(4) + '\t' + D.toPrecision(3) + '\t' + E.toPrecision(3) + '\t' + (F * divBy * conv).toExponential(4) + '\t' + G.toPrecision(3) + '\t' + H.toPrecision(3)
copyTextToClipboard(cText)
}
amUpdating = false
let cText = (conv * A / divBy).toPrecision(4) + '\t' + (conv * B / divBy).toPrecision(4)
if (Math.abs(C) > 0.001) cText += '\t' + (conv * C / divBy).toPrecision(4)
if (Math.abs(D) > 0.001) cText += '\t' + (conv * D / divBy).toPrecision(4)
if (Math.abs(E) > 0.001) cText += '\t' + (conv * E / divBy).toPrecision(4)
if (!Model.includes("Type")) copyTextToClipboard(cText)
let sText = "A: " + (conv * A / divBy).toPrecision(4) + ' B: ' + (conv * B / divBy).toPrecision(4)
if (Math.abs(C) > 0.001) sText += ' C: ' + (conv * C / divBy).toPrecision(4)
if (Math.abs(D) > 0.001) sText += ' D: ' + (conv * D / divBy).toPrecision(4)
if (Math.abs(E) > 0.001) sText += ' E: ' + (conv * E / divBy).toPrecision(4)
if (Model.includes("File") && fitData.length > 3) sText += ' -B/C:' + (-B / C).toPrecision(3)
if (!Model.includes("Type")) { Parameters = sText }
// conv = 1
ModelLabel = "Model Parameters"
if (Model.includes("File") && !Model.includes("Type") && fitData.length > 3) {
ModelLabel = "Model Parameters kg/mol"
const VPRT = isNaN(AA) ? "-" : Math.pow(10, AA - AB / (AC + T)) / 760 * 100000 / (8.312 * (T + 273.15))
const Gs2 = divBy / (conv * VPRT * A)
const G22 = conv * B / divBy
const VSA = -Area * 1e-20 * 6.02e23 / G22 / 1000 //Decided to do it /g instead of /kg
if (VPRT == "-") {
Values = "STSA = " + VSA.toPrecision(3) + " m²/g"
} else {
Values = "Gs2° = " + Gs2.toPrecision(4) + " m³/kg, G22°/v° = " + G22.toPrecision(4) + " kg/mol, STSSA = " + VSA.toPrecision(3) + " m²/g, c2_sat = " + VPRT.toPrecision(3) + " mol/m³"
}
}
G22conv = conv
newFit = false
}
} else {
// document.getElementById('Slideamax').style.visibility = "visible"
lastFit = ""; divBy = 1
}
for (let a2 = a2inc; a2 <= Math.min(amax, xmax + 0.05); a2 += a2inc) {
if (!Model.includes("Type")) {
n = a2 / (A - E * Math.pow(a2, 4) / 4 - D * Math.pow(a2, 3) / 3 - C * Math.pow(a2, 2) / 2 - B * a2)
G22 = E * a2 * a2 * a2 + D * a2 * a2 + C * a2 + B
} else {
if (Model.endsWith("Type IV")) {
n = A * B * a2 / (1 + B * a2) + C * (D * E * Math.pow(a2, D)) / ((1 + E * Math.pow(a2, D)))
G22 = C * D * D * E * Math.pow(a2, D - 1) / (E * Math.pow(a2, D) + 1) - C * D * D * E * E * Math.pow(a2, 2 * D - 1) / Math.pow(E * Math.pow(a2, D) + 1, 2) + A * B / (B * a2 + 1) - A * B * B * a2 / Math.pow(B * a2 + 1, 2)
}
if (Model.endsWith("Type V")) {
n = A * (B * a2 + 2 * E * Math.pow(a2, 2) + D * C * Math.pow(a2, D)) / (1 + B * a2 + E * Math.pow(a2, 2) + C * Math.pow(a2, D))
G22 = -1 / A + D * (D - 1) / A * C * (1 + B * a2 + C * Math.pow(a2, D)) * Math.pow(a2, D - 2) / Math.pow(B + D * C * Math.pow(a2, D - 1), 2)
}
if (Model.endsWith("Type VI")) {
n = A * B * a2 / (1 + B * a2) + C * (D * E * Math.pow(a2, D)) / ((1 + E * Math.pow(a2, D))) + F * (G * H * Math.pow(a2, G)) / ((1 + H * Math.pow(a2, G)))
G22 = F * G * G * H * Math.pow(a2, G - 1) / (H * Math.pow(a2, G) + 1) - F * G * G * H * H * Math.pow(a2, 2 * G - 1) / Math.pow(H * Math.pow(a2, G) + 1, 2) + C * D * D * E * Math.pow(a2, D - 1) / (E * Math.pow(a2, D) + 1) - C * D * D * E * E * Math.pow(a2, 2 * D - 1) / Math.pow(E * Math.pow(a2, D) + 1, 2) + A * B / (B * a2 + 1) - A * B * B * a2 / Math.pow(B * a2 + 1, 2)
}
}
if (n > 1 || n < -0.02) break
ICurve.push({ x: a2, y: Math.max(0, n * divBy) })
GCurve.push({ x: a2, y: G22conv * G22 / divBy })
NCurve.push({ x: a2, y: G22conv * G22 * n * divBy })
if (a2 > 0.025) naCurve.push({ x: a2, y: Math.max(0, n * divBy) / a2 })
if (n * divBy > 0.025) anCurve.push({ x: a2, y: a2 / (n * divBy) })
}
//Rounding error issues for the plot
n = amax / (A - E * Math.pow(amax, 4) / 4 - D * Math.pow(amax, 3) / 3 - C * Math.pow(amax, 2) / 2 - B * amax)
if (!Model.includes("Type")) {
n = amax / (A - E * Math.pow(amax, 4) / 4 - D * Math.pow(amax, 3) / 3 - C * Math.pow(amax, 2) / 2 - B * amax)
} else {
if (Model.endsWith("Type IV")) {
n = A * B * amax / (1 + B * amax) + C * (D * E * Math.pow(amax, D)) / ((1 + E * Math.pow(amax, D)))
}
if (Model.endsWith("Type V")) {
n = A * (B * amax + + 2 * E * Math.pow(amax, 2) + D * C * Math.pow(amax, D)) / (1 + B * amax + E * Math.pow(amax, 2) + C * Math.pow(amax, D))
}
if (Model.endsWith("Type VI")) {
n = A * B * amax / (1 + B * amax) + C * (D * E * Math.pow(amax, D)) / ((1 + E * Math.pow(amax, D))) + F * (G * H * Math.pow(amax, G)) / ((1 + H * Math.pow(amax, G)))
}
}
if (xmax + 0.05 >= amax && n <= 1 && n > 0) ICurve.push({ x: amax, y: n * divBy })
const dualPlot = false //Remnant of old code
let plotData = dualPlot ? [PCurve, ICurve] : [ICurve]
let lineLabels = dualPlot ? [Model, "Stat Therm"] : ["Stat Therm"]
let theColors = dualPlot ? ['#afd8ff', '#edc240'] : ['#edc240']
let thenaColors = ['#edc240', '#afd8ff']
let showLines = [true, true], showPoints = [0, 0]
let naData = [naCurve, anCurve]
let naLineLabels = ["⟨n2⟩/a2", "a2/⟨n2⟩"]
let G2y2Label = "a2/⟨n2⟩& "
let G2yAxisL1R2 = [1, 2]
if (Model.includes("File") && fitData.length > 3) {
plotData = [scaleData, ICurve]
lineLabels = ["Raw", "⟨n2⟩/a2", "a2/⟨n2⟩"]
theColors = ['#6f98ff', '#edc240']
thenaColors = ['#6f98ff', '#edc240', '#afd8ff']
showLines = [false, true, true], showPoints = [2, 0, 0]
naData = [naFitData, naCurve, anCurve]
naLineLabels = ["Raw", "⟨n2⟩/a2", "a2/⟨n2⟩"]
G2yAxisL1R2 = [1, 1, 2]
}
//Now set up all the graphing data detail by detail.
const prmap = {
plotData: plotData, //An array of 1 or more datasets
lineLabels: lineLabels, //An array of labels for each dataset
colors: theColors,
showLines: showLines,
showPoints: showPoints,
xLabel: "a2& ", //Label for the x axis, with an & to separate the units
yLabel: "⟨n2⟩&" + Units, //Label for the y axis, with an & to separate the units
y2Label: null, //Label for the y2 axis, null if not needed
yAxisL1R2: [], //Array to say which axis each dataset goes on. Blank=Left=1
logX: false, //Is the x-axis in log form?
xTicks: undefined, //We can define a tick function if we're being fancy
logY: false, //Is the y-axis in log form?
yTicks: undefined, //We can define a tick function if we're being fancy
legendPosition: 'top', //Where we want the legend - top, bottom, left, right
xMinMax: [,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
yMinMax: [,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
y2MinMax: [,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
xSigFigs: 'F2', //These are the sig figs for the Tooltip readout. A wide choice!
ySigFigs: 'F3', //F for Fixed, P for Precision, E for exponential
};
let ShowUnits = " "
if (Model.includes("File") && !Model.includes("Type") && fitData.length > 3) ShowUnits = "kg/mol"
const prmap1 = {
plotData: [GCurve, NCurve], //An array of 1 or more datasets
lineLabels: ["G22 / v", "N22"], //An array of labels for each dataset
xLabel: "a2& ", //Label for the x axis, with an & to separate the units
yLabel: "G22 / v&" + ShowUnits, //Label for the y axis, with an & to separate the units
y2Label: "N22& ", //Label for the y2 axis, null if not needed
yAxisL1R2: [1, 2], //Array to say which axis each dataset goes on. Blank=Left=1
logX: false, //Is the x-axis in log form?
xTicks: undefined, //We can define a tick function if we're being fancy
logY: false, //Is the y-axis in log form?
yTicks: undefined, //We can define a tick function if we're being fancy
legendPosition: 'top', //Where we want the legend - top, bottom, left, right
xMinMax: [,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
yMinMax: [,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
y2MinMax: [,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
xSigFigs: 'P3', //These are the sig figs for the Tooltip readout. A wide choice!
ySigFigs: 'P3', //F for Fixed, P for Precision, E for exponential
};
const prmap2 = {
plotData: naData, //An array of 1 or more datasets
lineLabels: naLineLabels, //An array of labels for each dataset
colors: thenaColors,
showLines: showLines,
showPoints: showPoints,
xLabel: "a2& ", //Label for the x axis, with an & to separate the units
yLabel: "⟨n2⟩ / a2&" + Units, //Label for the y axis, with an & to separate the units
y2Label: G2y2Label, //Label for the y2 axis, null if not needed
yAxisL1R2: G2yAxisL1R2, //Array to say which axis each dataset goes on. Blank=Left=1
logX: false, //Is the x-axis in log form?
xTicks: undefined, //We can define a tick function if we're being fancy
logY: false, //Is the y-axis in log form?
yTicks: undefined, //We can define a tick function if we're being fancy
legendPosition: 'top', //Where we want the legend - top, bottom, left, right
xMinMax: [,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
yMinMax: [0,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
y2MinMax: [,], //Set min and max, e.g. [-10,100], leave one or both blank for auto
xSigFigs: 'P3', //These are the sig figs for the Tooltip readout. A wide choice!
ySigFigs: 'P3', //F for Fixed, P for Precision, E for exponential
};
//Now we return everything - text boxes, plot and the name of the canvas, which is 'canvas' for a single plot
return {
Parameters: Parameters,
Values: Values,
ModelLabel: ModelLabel,
plots: [prmap, prmap1, prmap2],
canvas: ['canvas', 'canvas1', 'canvas2'],
};
}
//The fitting routine
fminsearch = function (fun, Parm0, x, y, Opt) {
if (!Opt) {
Opt = {}
};
if (!Opt.maxIter) {
Opt.maxIter = 1000
};
if (!Opt.step) { // initial step is 1/100 of initial value (remember not to use zero in Parm0)
Opt.step = Parm0.map(function (p) {
return p / 100
});
Opt.step = Opt.step.map(function (si) {
if (si == 0) {
return 1
} else {
return si
}
}); // convert null steps into 1's
};
if (!Opt.objFun) {
Opt.objFun = function (y, yp) {
return y.map(function (yi, i) {
return Math.pow((yi - yp[i]), 2)
}).reduce(function (a, b) {
return a + b
})
}
} //SSD
var cloneVector = function (V) {
return V.map(function (v) {
return v
})
};
var P0 = cloneVector(Parm0),
P1 = cloneVector(Parm0);
var n = P0.length;
var step = Opt.step;
var funParm = function (P) {
return Opt.objFun(y, fun(x, P))
} //function (of Parameters) to minimize multi-univariate screening
let lastfP = 1e-19, newfP = 0, lasti = 0
const Q = Math.sqrt((Opt.maxIter - 2000) / 2000)
const eps = Math.pow(10, -3 - Q)
for (var i = 0; i < Opt.maxIter; i++) {
for (var j = 0; j < n; j++) { // take a step for each parameter
P1 = cloneVector(P0);
P1[j] += step[j];
if ((!Opt.noNeg || P1[j] > 0) && funParm(P1) < funParm(P0)) { // if parm value going in the righ direction
step[j] = 1.2 * step[j]; // then go a little faster
P0 = cloneVector(P1);
} else {
step[j] = -(0.5 * step[j]); // otherwiese reverse and go slower
}
}
//Stop trying too hard
newfP = funParm(P0)
if (i > 1000 && newfP < lastfP && Math.abs((newfP - lastfP) / lastfP) < eps) break
lastfP = newfP
lasti = i
}
//console.log(lasti)
return P0
};
function clearOldName() { //This is needed because otherwise you can't re-load the same file name!
Loading = true
document.getElementById('Load').value = ""
}
function handleFileSelect(evt) {
var f = evt.target.files[0];
if (f) {
var r = new FileReader();
r.onload = function (e) {
var data = e.target.result;
LoadData(data)
}
r.readAsText(f);
} else {
return;
}
}
//Load data from a chosen file
function LoadData(S) {
Papa.parse(S, {
download: false,
header: true,
skipEmptyLines: true,
complete: papaCompleteFn,
error: papaErrorFn
})
}
function papaErrorFn(error, file) {
console.log("Error:", error, file)
}
function papaCompleteFn() {
var theData = arguments[0]
fitData = []; scaleData = []; lastFit = "", newFit = true
if (theData.data.length < 3) return //Not a valid data file
let maxY = 0
for (i = 0; i < theData.data.length; i++) {
theRow = theData.data[i]
if (theRow.Item.toUpperCase() == "DATA") {
fitData.push({ x: theRow.a, y: theRow.n })
scaleData.push({ x: theRow.a, y: theRow.n })
let y = parseFloat(theRow.n)
if (y > maxY) maxY = y
}
}
if (maxY == 0) return //Invalid data
divBy = 1
if (maxY > 1) { //The general fitting is easier if scaled from ~0-1
if (maxY <= 10000) divBy = 10000
if (maxY <= 1000) divBy = 1000
if (maxY <= 100) divBy = 100
if (maxY <= 10) divBy = 10
for (i = 0; i < fitData.length; i++) {
fitData[i].y /= divBy
}
}
Loading = false
Main()
}
function fallbackCopyTextToClipboard(text) { //Necessary if async not in browser
var textArea = document.createElement("textarea");
textArea.value = text;
document.body.appendChild(textArea);
textArea.focus();
textArea.select();
var elmnt = document.getElementById("network");
elmnt.scrollIntoView();
try {
var successful = document.execCommand('copy');
var msg = successful ? 'successful' : 'unsuccessful';
} catch (err) {
}
document.body.removeChild(textArea);
}
function copyTextToClipboard(text) {
if (!navigator.clipboard) {
fallbackCopyTextToClipboard(text);
return;
}
navigator.clipboard.writeText(text).then(function () {
}, function (err) {
});
}
function GetDemo() {
clearOldName()
const Demo = document.getElementById('Demo').value
const KUF = [[0.006281615, 0.647], [0.008034289, 0.7], [0.010176332, 0.77], [0.012317519, 0.884], [0.014945846, 0.997], [0.017477429, 1.076], [0.019716558, 1.164], [0.022345398, 1.251], [0.024779553, 1.33], [0.027310623, 1.435], [0.029842035, 1.522], [0.03227602, 1.61], [0.03490486, 1.697], [0.037143989, 1.785], [0.039772486, 1.89], [0.042303898, 1.978], [0.04473754, 2.083], [0.047171525, 2.17], [0.049605167, 2.275], [0.052038466, 2.398], [0.054666621, 2.521], [0.057001979, 2.67], [0.059531851, 2.836], [0.062060694, 3.055], [0.064586799, 3.415], [0.067465458, 5.682], [0.077297111, 6.12], [0.080705203, 6.216], [0.08197108, 6.251], [0.08469752, 6.33], [0.087034761, 6.382], [0.089567201, 6.417], [0.092001528, 6.487], [0.094631224, 6.53], [0.097066235, 6.565], [0.099500905, 6.618], [0.101935745, 6.661]]
const AlPO4 = [[0, 0.12], [0.025, 5.29], [0.039, 6.94], [0.049, 8.35], [0.069, 10], [0.082, 11.18], [0.116, 13.06], [0.151, 15.41], [0.165, 15.41], [0.175, 17.76], [0.22, 20.82], [0.244, 26], [0.267, 33.06], [0.276, 42.71], [0.276, 46.71], [0.277, 52.12], [0.278, 58], [0.278, 64.12], [0.278, 74.24], [0.279, 84.35], [0.28, 94.94], [0.281, 106.71], [0.285, 119.88], [0.285, 132.12], [0.292, 146.47], [0.318, 166], [0.384, 183.88], [0.606, 188.59]]
const CO2V=[[0.004, 3.2], [0.006, 4.8], [0.008, 6.2], [0.025, 7.7], [0.059, 9.7], [0.085, 11.2], [0.11, 13.1], [0.135, 14.6], [0.159, 16], [0.185, 17.3], [0.209, 18.3], [0.236, 19.5], [0.26, 20], [0.28, 21], [0.305, 22.2], [0.322, 34.8], [0.351, 55.2], [0.375, 77.1], [0.402, 97.6], [0.423, 108.7], [0.45, 117.5], [0.476, 123.9], [0.504, 127.7], [0.529, 130.6], [0.555, 132.6], [0.579, 134.5], [0.605, 136.3], [0.631, 137.7], [0.655, 138.9], [0.681, 139.9], [0.705, 140.7], [0.731, 141.4], [0.756, 141.7], [0.78, 142.2], [0.806, 142.7], [0.831, 143.1], [0.856, 143.6], [0.88, 143.9], [0.905, 144.2], [0.932, 144.4], [0.956, 144.6], [0.981, 144.7]]
const PCN = [[0, 0], [0.001, 15.9], [0.002, 26.2], [0.003, 36.6], [0.005, 46.9], [0.007, 57.2], [0.03, 91.6], [0.049, 110.5], [0.075, 129.5], [0.095, 146.7], [0.112, 157], [0.132, 169], [0.152, 182.8], [0.172, 196.6], [0.192, 210.3], [0.213, 224.1], [0.231, 237.8], [0.253, 253.3], [0.272, 268.8], [0.293, 286], [0.314, 304.9], [0.336, 323.9], [0.357, 342.8], [0.383, 365.2], [0.402, 384.1], [0.425, 404.7], [0.443, 421.9], [0.465, 440.9], [0.482, 461.5], [0.502, 523.4], [0.524, 618.1], [0.56, 664.5], [0.577, 673.1], [0.596, 683.4], [0.608, 688.6], [0.631, 702.4], [0.65, 711], [0.67, 726.5], [0.69, 769.5], [0.71, 838.3], [0.731, 888.2], [0.754, 917.4], [0.768, 922.6], [0.801, 934.6], [0.817, 936.3], [0.85, 941.5], [0.874, 943.2], [0.895, 946.7], [0.899, 946.7], [0.922, 948.4], [0.956, 951.8], [0.961, 951.8], [0.985, 953.5]]
let theData = []
if (Demo == "KUF-1A NH3 IV") theData = KUF
if (Demo == "AlPO4 H2O IV") theData = AlPO4
if (Demo == "CO2 V") theData = CO2V
if (Demo == "PCN-53 CO2 VI") theData = PCN
lastFit=""
fitData = [];scaleData=[]
if (theData.length < 3) return
let maxY = 0
for (i = 0; i < theData.length; i++) {
fitData.push({ x: theData[i][0], y: theData[i][1] })
scaleData.push({ x: theData[i][0], y: theData[i][1] })
let y = theData[i][1]
if (y > maxY) maxY = y
}
if (maxY == 0) return //Invalid data
divBy = 1
if (maxY > 1) { //The general fitting is easier if scaled from ~0-1
if (maxY <= 10000) divBy = 10000
if (maxY <= 1000) divBy = 1000
if (maxY <= 100) divBy = 100
if (maxY <= 10) divBy = 10
for (i = 0; i < fitData.length; i++) {
fitData[i].y /= divBy
}
}
Loading = false
Main()
}
Background
The adsorption (onto) and absorption (into) of molecules such as water onto/into surfaces [the sorbate molecules interact with the sorbent] can be experimentally measured, and the curve of activity (very often just concentration) versus amount ad/ab-sorbed is called the isotherm. These isotherms have distinctive shapes, often with plateaux but sometimes with inflexion points. There has been a long history of describing these isotherms with models based on some plausible science of what is happening in, for example, monolayers and multilayers.
As happens so often in science, helpful abstractions based on simple, limiting cases gain the aura of providing fundamental insights into complex processes. So the various isotherm models have been used in cases where their fundamental assumptions simply cannot apply. At the same time, the complexities of real isotherms invite alternative models, with the official body IUPAC listing 80+ such models. The whole area is a mess because, as has often been shown1, it is possible to fit the same real-world isotherm data with very different isotherm models based on different assumptions.
Fortunately, by going back to the fundamentals, it's rather straightforward to produce an assumption-free theory that allows us to understand all basic sorption processes.
Based on the recently published papers1,2, we first look at how the theory gives us insightful, assumption-free curves for any isotherm, with very little effort. We then see how some modest assumptions behind two classes of isotherms can give us fits where the parameters have deep meanings3, more insightful than those which are tied to the numerous models based on inappropriate assumptions.
The Gs2 and G22 / v curves1,2
An isotherm is just a plot of 〈n2〉 (the brackets mean "ensemble average"), the amount sorbed per unit mass of sorbent, versus a2, the activity of the sorbate. The "2" means that this is the sorbate as opposed to "s" the surface. If, instead, we plot 〈n2〉/a2 versus a2, we end up with a plot that, when multiplied by a constant, gives us Gs2, as shown in the recent papers 1,2. This is a Kirkwood-Buff integral which tells us how much extra "2" we have on the surface compared to an average without sorption. Typically, it starts off high, meaning that there's a strong attraction, then it reduces because there are fewer available sites then it increases for interesting reasons discussed next.
If you take the derivative of the inverse curve, a2/〈n2〉 you directly obtain G22/v. This is a measure of how many sorbates you have together compared to the average, divided by the "interfacial volume" v which is discussed later. This a2/〈n2〉 typically starts off negative, you have fewer sorbates together than the average for the simple reason that if you have one sorbate somewhere you cannot have a second one in the same place; this is the "excluded volume" effect. Eventually you reach a point where you G22/v becomes positive - this means that you have net sorbate-sorbate interactions. The point at which this happens is exactly at the point where Gs2 starts to increase. Now you have more surface-sorbate interactions because sorbate-sorbate interactions (which are themselves enabled by the surface) mean that any sorbate at the surface brings another (fraction of a) sorbate with it.
This molecular picture applies to all possible sorption curves and makes no mention of "monolayer coverage" or "sorption sites" or "micropores" and is derived straight from the isotherm itself. What's not to like?
Cooperative Fits
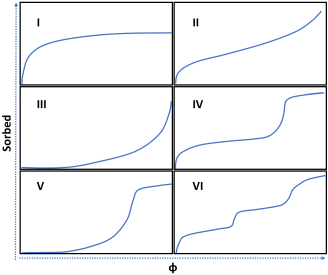 With sufficient number of parameters a master equation can replicate any isotherm, but loses meaning. An alternative is to make an approximation that the system is the combination of a simple curve and one or more mth order effects giving cooperative clustering and sigmoidal curves, as we propose here3. There are 3 cases defined by IUPAC, Type IV, Type V,Type VI. If you have obvious multiple steps, choose Type VI from the start, and obtain the 8 parameters. Types IV and V have similar shapes, so how to choose between them? Because Type V has 4 parameters, if the fit is OK then use it, otherwise go to Type IV and gain the benefit of 5 parameters. The equations used involve the scaling constants N, in mol/kg with the other parameters being dimensionless:
With sufficient number of parameters a master equation can replicate any isotherm, but loses meaning. An alternative is to make an approximation that the system is the combination of a simple curve and one or more mth order effects giving cooperative clustering and sigmoidal curves, as we propose here3. There are 3 cases defined by IUPAC, Type IV, Type V,Type VI. If you have obvious multiple steps, choose Type VI from the start, and obtain the 8 parameters. Types IV and V have similar shapes, so how to choose between them? Because Type V has 4 parameters, if the fit is OK then use it, otherwise go to Type IV and gain the benefit of 5 parameters. The equations used involve the scaling constants N, in mol/kg with the other parameters being dimensionless:
The Demo files are those used in the third paper3 which is about Types IV and V plus a Type V fit for reference. See below for how to load your own isotherm data into the app.
Type IV
`"〈"n_2"〉"=N_a(A_1a_2)/(1+A_1a_2)+N_b(mA_ma_2^m)/(1+A_ma_2^m)`
Note that although you can fit this equation directly, it is difficult to get to the optimum. The app fits to the parameter `ln(A_m)/m` and uses information from the data such as the height of the cooperative step, and the position and gradient of the steepest gradient of the step. Doing this, the fitting starts with good estimates so is faster and more reliable.
Type V
`"〈"n_2"〉"=N(A_1a_2+mA_ma_2^m)/(1+A_1a_2+A_ma_2^m)`
Type VI and Type VI-like
IUPAC define Type VI as "layer-by-layer adsorption on a highly uniform nonporous surface". But it is common to use Type VI for multi-step sorption on porous surfaces - rarely are they called "Type VI-like". The equation is equally valid for both types as it makes no assumptions about the reasons for the multiple steps.
`"〈"n_2"〉"=N_a(A_1a_2)/(1+A_1a_2)+N_b(mA_ma_2^m)/(1+A_ma_2^m)+N_c(nA_na_2^n)/(1+A_na_2^n)`
The key things to look for are the m values. If they are small (<~5) then it's saying that not much cooperative clustering is taking place, when they are large then that's a strong sign of such clustering. At the same time, large peaks in G22 are a good indication of strong cooperative clustering that coincides with the sort of sigmoidal curve for which the approach is best suited.
Analysis shows that fit quality can be similar for multiple pairs of Am and m values. This is partly a numeric issue and partly a data quality issue. If m is super-important then more data around the step is required to pin it down. But remember that m is intended to capture the broad essence of cluster sizes which will, in any case, cover a range of sizes, so it might not be worth the effort to obtain a more precise value.
App inputs
The Fit Quality is a trade-off parameter. A low value does a rapid fit but the data might fit poorly. A high value might give a better fit but take longer. Start with something like 3 and see if changing it makes a difference. Even with a high value, if the fit quality isn't changing significantly, the algorithm halts.
Many units are used for isotherms - choose the one closest to yours. The Sorbate choice is there in case you are specifying weight units, to allow conversion to molar values. The other values for each sorbate are used in other apps and are ignored here. Temperature is for calculation of `-RTln(A_m)` a key thermodynamic value.
Fitting your own data
A set of representative isotherms based on the different models is provided in Cooperative-Isotherms.zip. The individual isotherms are in .csv format with the first row Item, a, n and subsequent rows with Data, a_value, n_value. This slightly unfamiliar format reflects modern JavaScript file handling methods. The "a" values are short for a2
Prepare your own file in that format. The n values should be in one of the unit choices offered by the app.
Seishi Shimizu and Nobuyuki Matubayasi, Sorption: A Statistical Thermodynamic Fluctuation Theory, Langmuir 2021, 37, 24, 7380–7391
Seishi Shimizu and Nobuyuki Matubayasi, Cooperative Sorption on Porous Materials, Langmuir 2021, 37, 34, 10279–10290
Olivia P. L. Dalby, Steven Abbott, Nobuyuki Matubayasi, and Seishi Shimizu, Cooperative Sorption on Heterogeneous Surfaces, Langmuir 2022, https://doi.org/10.1021/acs.langmuir.2c01750


